GO is currently deprecating our public SPARQL endpoint, with plans to deprecate all related products in the future. Blazegraph journals are available here for advanced users who do not need support. If you have questions, please contact us through the GO Helpdesk
Introduction
Gene Ontology is developing Ontologies and Frameworks to help annotate biology in a consistent way and help users to integrate and reuse our data in their approaches. The Gene Ontology Causal Activity Model (or GO-CAM) recently published allows to curate knowledge in a much more expressive and traceable way and extend the querying capabilities of the standard GO annotations. [ Learn more about GO-CAMs ]
To support the creation, storage and querying of GO-CAMs, GO is relying on RDF / OWL, a directed, labeled graph data format to represent rich and complex knowledge. RDF graphs can be queried and manipulated with SPARQL, one of the standards of the Semantic Web.
In the following document, we present some notions of RDF, OWL and SPARQL to get an understanding of how the GO SPARQL endpoint is structured and how to create queries to retrieve specific pieces of knowledge.
Resource Description Framework (RDF)
The core structure of RDF is a triple consisting of a subject, a predicate (a relationship) and an object. A RDF graph is a set of such triples chaining different pieces of knowledge across multiple subjects and objects.
` [ Subject ] — predicate —> [ Object ] `
There are three kinds of nodes in a RDF graph:
- IRIs: a generalization of URIs that permits a wider range of Unicode characters. They are used to uniquely describe an object over the web and as such should be permanent URL, or PURL. As an example, the GO term
defense response to Gram-negative bacteriumis uniquely and permanently defined on the web by the PURLhttps://purl.obolibrary.org/obo/GO_0050829. - Literals: used to store values describing an object (e.g. strings, numbers, dates). For instance, one can record that the gene with id
P02340has for label the literalCellular tumor antigen p53with the following triple :<https://www.uniprot.org/uniprot/P02340> rdfs:label "Cellular tumor antigen p53". - Blank nodes: local identifiers always scoped to an RDF store that are not persistent. In the GO RDF, blank nodes starts with an underscore, e.g.
_:t432889.
[ Learn more about RDF graph ]
Web Ontology Language (OWL)
OWL and OWL2 are ontology languages for the Semantic Web which allow to describe ontology components such as classes, properties, individuals and data values. Any OWL2 ontology can also be viewed as an RDF graph. OWL can be reasoned with by computer programs both to check the consistency of the aggregated knowledge (GO is using Shex to ensure the validity of GO-CAMs) and to make implicit knowledge explicit (e.g. if A is a B and B is part of C, then we can explicitly state that A is part of C).
[ Learn more about OWL2 ] [ Learn more about ontology relations in GO ]
In addition of using OWL to describe ontologies, GO-CAMs rely on the Relation Ontology (RO) and Basic Formal Ontology (BFO) to describe the relations between genes, molecular function, biological process and cellular component. In the following example, the GO-CAM informs that the activity sequence-specific DNA binding enabled by the IRF3 Hsap gene directly positively regulates the DNA-binding transcription activator activity of IRF3 Hsap, and that both activities occurs in the nucleus:

In this other GO-CAM, one can read that the DNA-binding transcription factor activity enabled by zip-2 Cele is causally upstream of, positive effect of two molecular activities: the deaminase activity on the left and an unknown activity on the right generically called Molecular function but which is enabled by irg-2 Cele. In addition, all those activities are described as being part of the defense response to Gram-negative bacterium biological process.
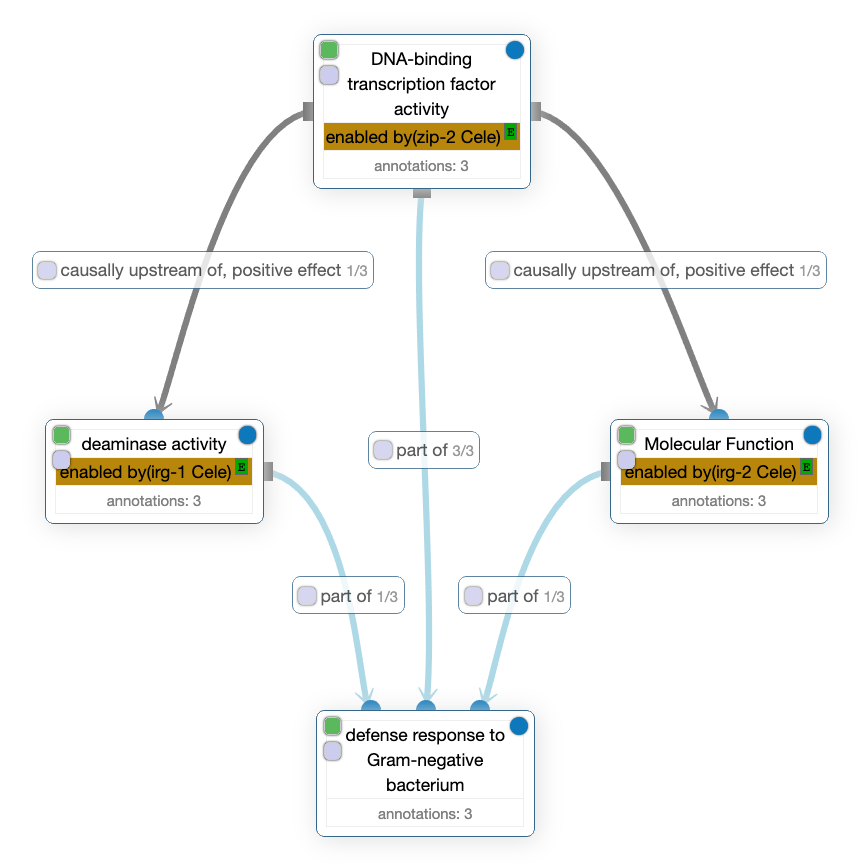
In those GO-CAM, the predicates enabled by, directly positively regulates and causally upstream of, positive effect comes from RO while occurs in and part of comes from BFO. Here is a table summarizing those predicates and their IRIs, used to uniquely identify them on the web:
| Relation / Predicate | Description | IRI |
|---|---|---|
| enabled by | Description | http://purl.obolibrary.org/obo/RO_0002333 |
| directly positively regulates | Description | http://purl.obolibrary.org/obo/RO_0002629 |
| causally upstream of, positive effect | Description | http://purl.obolibrary.org/obo/RO_0002304 |
| occurs in | Description | http://purl.obolibrary.org/obo/BFO_0000066 |
| part of | Description | http://purl.obolibrary.org/obo/BFO_0000050 |
The second model can be visualized as a graph here and is available as a Turtle/TTL file here. Let’s take a look at a sample of that TTL file:
<http://model.geneontology.org/5b91dbd100001993/5b91dbd100001995> <http://purl.org/pav/providedBy>
"http://www.wormbase.org"^^<http://www.w3.org/2001/XMLSchema#string> ;
<http://purl.obolibrary.org/obo/BFO_0000050> <http://model.geneontology.org/5b91dbd100001993/5b91dbd100001996> ;
<http://purl.obolibrary.org/obo/RO_0002333> <http://model.geneontology.org/5b91dbd100001993/5b91dbd100001994> ;
<http://purl.obolibrary.org/obo/RO_0002304> <http://model.geneontology.org/5b91dbd100001993/5b91dbd100002002> ,
A few notes about the sugar syntaxes of Turtle/TTL files:
;is used for predicate lists to avoid rewriting the same subject at the beginning of multiple lines/triples,is used for object lists in a same way but to avoid rewriting both the same subject and the same predicate at the beginning of multiple lines/triplesais a shortcut for therdf:typepredicate, and reads asis of typeoris a
With that syntax in mind, the above triples can be read as:
- the subject
<http://model.geneontology.org/5b91dbd100001993/5b91dbd100001995>isprovided by(<http://purl.org/pav/providedBy>) wormbase AND - it is
part of(<http://purl.obolibrary.org/obo/BFO_0000050>) the object<http://model.geneontology.org/5b91dbd100001993/5b91dbd100001996>AND - it is
enabled by(<http://purl.obolibrary.org/obo/RO_0002333>) this other object<http://model.geneontology.org/5b91dbd100001993/5b91dbd100001994>AND - it is
causally upstream of, positive effect(<http://purl.obolibrary.org/obo/RO_0002304>) of this third object<http://model.geneontology.org/5b91dbd100001993/5b91dbd100002002>
In other parts of the Turtle/TTL file, one can also see additional triples relating to these subjects and objects:
<http://model.geneontology.org/5b91dbd100001993/5b91dbd100001994> <http://purl.org/pav/providedBy> "http://www.wormbase.org"^^<http://www.w3.org/2001/XMLSchema#string> ;
a <http://www.w3.org/2002/07/owl#NamedIndividual> , <http://identifiers.org/wormbase/WBGene00019327> ;
...
<http://model.geneontology.org/5b91dbd100001993/5b91dbd100001995> <http://purl.org/pav/providedBy> "http://www.wormbase.org"^^<http://www.w3.org/2001/XMLSchema#string> ;
a <http://www.w3.org/2002/07/owl#NamedIndividual> , <http://purl.obolibrary.org/obo/GO_0003700> ;
...
<http://model.geneontology.org/5b91dbd100001993/5b91dbd100001996> <http://purl.org/pav/providedBy> "http://www.wormbase.org"^^<http://www.w3.org/2001/XMLSchema#string> ;
a <http://www.w3.org/2002/07/owl#NamedIndividual> , <http://purl.obolibrary.org/obo/GO_0050829> ;
...
<http://model.geneontology.org/5b91dbd100001993/5b91dbd100002002> <http://purl.org/pav/providedBy> "http://www.wormbase.org"^^<http://www.w3.org/2001/XMLSchema#string> ;
a <http://www.w3.org/2002/07/owl#NamedIndividual> , <http://purl.obolibrary.org/obo/GO_0003674> ;
Which reads as:
- the subject
<http://model.geneontology.org/5b91dbd100001993/5b91dbd100001994>is an individual (<http://www.w3.org/2002/07/owl#NamedIndividual>) and an instance of the genezip-2(<http://identifiers.org/wormbase/WBGene00019327>) - the subject
<http://model.geneontology.org/5b91dbd100001993/5b91dbd100001995>is an individual (<http://www.w3.org/2002/07/owl#NamedIndividual>) and an instance of the GO termDNA-binding transcription factor activity(<http://purl.obolibrary.org/obo/GO_0003700>) - the subject
<http://model.geneontology.org/5b91dbd100001993/5b91dbd100001996>is an individual (<http://www.w3.org/2002/07/owl#NamedIndividual>) and an instance of the GO termdefense response to Gram-negative bacterium(<http://purl.obolibrary.org/obo/GO_0050829>) - the subject
<http://model.geneontology.org/5b91dbd100001993/5b91dbd100002002>is an individual (<http://www.w3.org/2002/07/owl#NamedIndividual>) and an instance of the GO termmolecular function(<http://purl.obolibrary.org/obo/GO_0003674>)
Together, the first block of our Turtle/TTL file can now be translated to:
This instance of the DNA-binding transcription factor activity provided by wormbase
is part of the defense response to Gram-negative bacterium AND
is enabled by the gene zip-2 AND
is causally upstream of, positive effect of a molecular function
This introduction was important to understand the underlying RDF/OWL structure used to store GO-CAMs. In the next section, we’ll see how to query the GO SPARQL endpoint to retrieve specific pieces of knowledge.
SPARQL to query GO-CAMs
SPARQL is the query language for RDF graphs. A number of scientific databases are adopting RDF/OWL and SPARQL for their consistent way to describe and link complex knowledge. The most simple query that can be done in SPARQL is to request a list of all triples:
SELECT * WHERE {
?sub ?pred ?obj .
}
LIMIT 100
OFFSET 0
Similar to SQL, SPARQL uses the SELECT keyword to define the fields that will be returned in the response. * means all the fields who have been defined in the WHERE block, in this example: ?sub, ?pred and ?obj. Since there is no constraint on either the value of the subject, the predicate or the object, this query will return all the triples, however the LIMIT parameter will limit the response to the first 100 triples. The OFFSETparameter was introduced here to illustrate how pagination can be done by skipping the first x triples.
The GO RDF store is a quad store that contains multiple graphs. Each GO-CAM is for instance stored as a subgraph in the GO RDF store, allowing to group the relevant triples together and perform a simple query to retrieve them:
SELECT * WHERE {
GRAPH <http://model.geneontology.org/5b91dbd100001993> {
?sub ?pred ?obj .
}
}
To simplify the writing and readability of those queries, SPARQL offers a PREFIX keyword which defines an alias to a base IRI. The above query can for instance also be written as:
PREFIX metago: <http://model.geneontology.org/>
SELECT * WHERE {
GRAPH metago:5b91dbd100001993 {
?sub ?pred ?obj .
}
}
Let’s dive into more useful queries and retrieve for instance all the genes contained in our previous GO-CAM:
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs:<http://www.w3.org/2000/01/rdf-schema#>
PREFIX owl: <http://www.w3.org/2002/07/owl#>
PREFIX metago: <http://model.geneontology.org/>
PREFIX enabled_by: <http://purl.obolibrary.org/obo/RO_0002333>
SELECT distinct ?identifier ?name
WHERE {
GRAPH metago:5b91dbd100001993 {
?s enabled_by: ?gpnode .
?gpnode rdf:type ?identifier .
}
?identifier rdfs:label ?name
}
Here, we retrieve all the enablers (genes) of the GO-CAM metago:5b91dbd100001993. We then retrieve their identifier (IRIs) and retrieve their labels name (gene names). Note that the rdfs:label predicate used to retrieve the subject label is written outside the GRAPH block, as the gene information is shared by all GO-CAMs and is described outside of that GO-CAM subgraph.
In the same way, one can retrieve the list of GO terms in a given GO-CAM:
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX metago: <http://model.geneontology.org/>
PREFIX owl: <http://www.w3.org/2002/07/owl#>
PREFIX definition: <http://purl.obolibrary.org/obo/IAO_0000115>
PREFIX BP: <http://purl.obolibrary.org/obo/GO_0008150>
PREFIX MF: <http://purl.obolibrary.org/obo/GO_0003674>
PREFIX CC: <http://purl.obolibrary.org/obo/GO_0005575>
SELECT distinct ?goaspect ?goids ?gonames ?definitions WHERE {
GRAPH metago:5b91dbd100001993 {
?entity rdf:type owl:NamedIndividual .
?entity rdf:type ?goids
}
VALUES ?goaspect { BP: MF: CC: } .
?goids rdfs:subClassOf+ ?goaspect .
?goids rdfs:label ?gonames .
?goids definition: ?definitions .
}
ORDER BY DESC(?gocam)
Here the code gets a little more complicated. First, notice the larger use of PREFIX which simplifies the readability of the actual query contained in the SELECT block. The SELECT line is used to display the values of the fields we want to retrieve, here the GO terms aspect ?goaspect, id goids, name gonames and definition definitions. The distinct keyword is used to ensure that a GO term would only be shown once in the response. On the contrary, if you want to retrieve all the instances of each GO term, the distinct keyword would have to be removed.
In the GRAPH block, we retrieve all the entities that are individuals, then we store their ids in goids. With the VALUES keyword, we create a list containing the root BP, MF and CC terms. We then filter every goids previously identified by ensuring they are a subclass of one of those GO root terms. Indeed, some of those individuals could be instances of genes or instances of other ontology terms used in the GO-CAM. As an example, GO-CAMs can also contain UBERON and CL ontology terms. From that list of goids, we then retrieve their labels with rdfs:label and their definition with the PREFIX definition: which is an alias to <http://purl.obolibrary.org/obo/IAO_0000115>.
A useful query also consist in retrieving the list of all GO-CAMs and to annotate them with some information. The following query retrieves for example, the list of all GO-CAMs with their title and date:
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX metago: <http://model.geneontology.org/>
PREFIX dc: <http://purl.org/dc/elements/1.1/>
SELECT ?gocam ?date ?title WHERE {
GRAPH ?gocam {
?gocam metago:graphType metago:noctuaCam .
?gocam dc:title ?title ;
dc:date ?date .
}
}
ORDER BY DESC(?date)
The SELECT line indicates we’ll be retrieving the gocam id, its date and its title. Note the GRAPH ?gocam where gocam is not constrained. This means SPARQL will be iterating through all graphs and assigned each graph to a variable ?gocam. Next, we filter the graphs retrieved by stating they must be of type metago:noctuaCam (type of a GO-CAM graph). The line after, we retrieve the title dc:title and the date dc:date of the current GO-CAM. At the end, SPARQL is asked to order all results with the value of the field ?date.
One can refine the previous query to retrieve all the GO-CAMs containing a given gene (e.g. Sox2 / MGI:98364):
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX metago: <http://model.geneontology.org/>
PREFIX enabled_by: <http://purl.obolibrary.org/obo/RO_0002333>
SELECT distinct ?gocam WHERE {
GRAPH ?gocam {
?gocam metago:graphType metago:noctuaCam .
?entity enabled_by: ?gpnode .
?gpnode rdf:type ?identifier .
FILTER( ?identifier = <http://identifiers.org/mgi/MGI:98364> ) .
}
}
The operator FILTER will keep only the gocams (?gocam) who have the requested gene id (?identifier).
This type of query can also be refined to retrieve all the GO-CAMs containing a given GO term (e.g. neuron apoptotic process / GO:0051402):
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX metago: <http://model.geneontology.org/>
PREFIX owl: <http://www.w3.org/2002/07/owl#>
SELECT distinct ?gocam WHERE {
GRAPH ?gocam {
?gocam metago:graphType metago:noctuaCam .
?entity rdf:type owl:NamedIndividual .
?entity rdf:type ?identifier .
FILTER( ?identifier = <http://purl.obolibrary.org/obo/GO_0051402> )
}
}
Federated Queries
Lastly, Federated Queries. Those specific types of queries are designed to retrieve specific pieces of information from multiple SPARQL endpoints (e.g. sparql.uniprot.org, sparql.wikipathway.org, etc). The concept of federated queries rely on the selection and sharing of unique IRIs to describe in a consistent way each entity across RDF stores. If a gene is universally identified by an IRI, then it one can request data about that gene across multiple databases in a single query. This technique, albeit powerful can however suffer from latency and can fail if any of the endpoint is unavailable. If you are mapping IDs between different endpoints, it’s critical to ensure you are gathering data about the same entity. [ Learn more about Federated Queries ].